

import numpy as np
# Scalars
# Definition: A single number (0D), e.g., a learning rate in AI models
learning_rate = 0.01 # Scalar for gradient descent step size
print("Scalar (Learning Rate):", learning_rate)
# Real-world: Controls how fast a neural network learns
# Operation: Simple arithmetic
scaled_value = learning_rate * 100
print("Scaled Scalar:", scaled_value)
# Vectors
# Definition: 1D array, e.g., feature vector in machine learning
feature_vector = np.array([0.5, 0.8, 0.2]) # Represents a data point (e.g., customer purchase history)
print("\nVector (Feature Vector):", feature_vector)
# Real-world: Used in recommendation systems or word embeddings
# Operation: Dot product (measures similarity)
another_vector = np.array([0.1, 0.4, 0.7])
dot_product = np.dot(feature_vector, another_vector)
print("Dot Product:", dot_product)
# Matrices
# Definition: 2D array, e.g., for image data or linear transformations
image_patch = np.array([[1, 2, 3], [4, 5, 6]]) # 2x3 matrix (e.g., grayscale image patch)
print("\nMatrix (Image Patch):\n", image_patch)
# Real-world: Used in CNNs for image processing
# Operation: Matrix multiplication (e.g., for transformations)
weights = np.array([[0.1, 0.2], [0.3, 0.4], [0.5, 0.6]]) # 3x2 weight matrix
result = np.matmul(image_patch, weights) # 2x2 result
print("Matrix Multiplication Result:\n", result)
# Tensors
# Definition: nD array (n≥0), e.g., for RGB images or video data
rgb_image = np.random.rand(224, 224, 3) # 3D tensor (height, width, RGB channels)
print("\nTensor (RGB Image Shape):", rgb_image.shape)
# Real-world: Input to CNNs like ResNet for image classification
# Operation: Tensor slicing (e.g., extract red channel)
red_channel = rgb_image[:, :, 0] # First channel (R)
print("Red Channel Shape:", red_channel.shape)
# Example: 4D tensor for a batch of images
batch_images = np.random.rand(2, 4, 4, 3) # Batch of 2 RGB 4x4 images
print("Batch Tensor:", batch_images)
Scalar (Learning Rate): 0.01 Scaled Scalar: 1.0 Vector (Feature Vector): [0.5 0.8 0.2] Dot Product: 0.51 Matrix (Image Patch): [[1 2 3] [4 5 6]] Matrix Multiplication Result: [[2.2 2.8] [4.9 6.4]] Tensor (RGB Image Shape): (224, 224, 3) Red Channel Shape: (224, 224) Batch Tensor Shape: [[[[0.04087357 0.57134536 0.32519568] [0.34167714 0.62335238 0.46628049] [0.07720674 0.77133272 0.36870046] [0.72117031 0.04701737 0.86162155]] [[0.85597285 0.14884848 0.48867849] [0.85831712 0.9510796 0.58200469] [0.57347744 0.24808471 0.41284116] [0.8178081 0.97333519 0.10092528]] [[0.04708062 0.04044213 0.22104739] [0.08955609 0.82080325 0.11358177] [0.91882124 0.60249004 0.72110625] [0.12282592 0.6878657 0.75100109]] [[0.79111113 0.02168534 0.89053718] [0.62379676 0.28582163 0.36874591] [0.29939015 0.49078382 0.96479222] [0.9737543 0.02131901 0.03780112]]] [[[0.89632584 0.97437265 0.16876449] [0.64941646 0.63884824 0.98995114] [0.44964252 0.72292501 0.44936663] [0.7425726 0.38513847 0.28983086]] [[0.31092603 0.88805527 0.1764936 ] [0.16567742 0.49115005 0.49396291] [0.01596826 0.98731473 0.70769769] [0.25895077 0.37736448 0.73386227]] [[0.09645565 0.36557884 0.39174251] [0.79035446 0.33778424 0.47940533] [0.64473504 0.038141 0.17750133] [0.78799436 0.35127631 0.31383826]] [[0.02208777 0.60393952 0.44482964] [0.29335764 0.16871938 0.34279193] [0.9368042 0.80870572 0.74452877] [0.67187876 0.11662868 0.86089061]]]]
In AI, scalars are used to represent constants, weights, or scaling factors.
In AI, vectors represent data points, features, or weights.
used to represent linear transformations or datasets in AI.
++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++ ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
Matrix: A rectangular array of numbers. Example:
$$ A = \begin{bmatrix} 1 & 2 \\ 3 & 4 \end{bmatrix} \in \mathbb{R}^{2 \times 2} $$
Dimensions: Rows × Columns
Addition/Subtraction: Element-wise (same dimensions)
Scalar Multiplication:
$$ \alpha A = \begin{bmatrix} \alpha a_{11} & \alpha a_{12} \\ \alpha a_{21} & \alpha a_{22} \end{bmatrix} $$
Matrix Multiplication:
$$ C = A \cdot B \quad \text{(valid only if columns of A = rows of B)} $$
Transpose:
$$ A^T = \text{flip rows and columns} $$
Identity Matrix:
$$ I_n = \text{square matrix with 1s on diagonal} $$
| Type | Property Example |
|---|---|
| Square | Same number of rows and columns |
| Diagonal | Non-zero entries only on diagonal |
| Symmetric | $A = A^T$ |
| Orthogonal | $A^T A = I$ (columns are orthonormal) |
| Zero Matrix | All elements are zero |
For $A \in \mathbb{R}^{n \times n}$, $A^{-1}$ satisfies:
$$ A A^{-1} = A^{-1} A = I $$
Only exists if $\det(A) \neq 0$ and $A$ is square.
Scalar value that can be computed from a square matrix:
$$ \text{det}(A) $$
Used to check invertibility and volume scaling.
Sum of diagonal elements of a square matrix:
$$ \text{tr}(A) = \sum_i a_{ii} $$
For $A \vec{v} = \lambda \vec{v}$:
| Type | Form | Use Case |
|---|---|---|
| LU | $A = LU$ | Solving systems |
| QR | $A = QR$ | Least squares, orthogonal bases |
| SVD | $A = U \Sigma V^T$ | PCA, compression, LSA |
| Eigendecomp | $A = V \Lambda V^{-1}$ | PCA, diagonalization |
| Matrix Concept | Application |
|---|---|
| Multiplication | Neural networks, transformations |
| Transpose | Covariance, dot product |
| Inverse | Solving linear systems |
| Rank | Dimensionality and redundancy |
| SVD/PCA | Dimensionality reduction |
A matrix norm measures the "size", "length", or "magnitude" of a matrix — similar to how a vector norm measures a vector’s length.
Think of it as: “How much can this matrix stretch or shrink a vector?”
| Norm | Definition | Interpretation | Example/Notes | ||
|---|---|---|---|---|---|
| Frobenius Norm $|A|_F$ | $\sqrt{ \sum_{i,j} a_{ij}^2 }$ | Like Euclidean norm for matrices | Used a lot in ML | ||
| 1-Norm $|A|_1$ | Max absolute column sum | Worst-case vertical stretching | $\max_j \sum_i | a_{ij} | $ |
| Infinity Norm $|A|_\infty$ | Max absolute row sum | Worst-case horizontal stretching | $\max_i \sum_j | a_{ij} | $ |
| 2-Norm (Spectral Norm) $|A|_2$ | Largest singular value of $A$ | Max stretching factor along any direction | Related to principal component, SVD, etc. |
The condition number tells you how sensitive a system or a matrix operation is to small changes in input.
In ML and numerical algorithms, lower condition numbers = more stable, reliable results.
For a non-singular matrix $A$:
$$ \text{cond}(A) = \|A\| \cdot \|A^{-1}\| $$For 2-norm:
$$ \text{cond}_2(A) = \frac{\sigma_{\text{max}}}{\sigma_{\text{min}}} $$Where $\sigma$ = singular values from SVD.
| Use Case | Impact |
|---|---|
| Solving Linear Systems | High condition number → errors get amplified |
| Inverting Matrices | Poor conditioning → instability |
| Gradient Descent | Ill-conditioned Hessian → slow convergence |
| Deep Learning | Can cause vanishing/exploding gradients |
++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++ ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
When a matrix doesn’t have an inverse (like when it’s non-square or rank-deficient), we use the pseudoinverse instead.
The Moore-Penrose Pseudoinverse $A^+$ of a matrix $A$ is a generalization of the inverse:
If $A$ is a $m \times n$ matrix, then $A^+$ is a $n \times m$ matrix.
| Scenario | Why use Pseudoinverse |
|---|---|
| $A$ is not square | So no usual inverse exists |
| $A$ is not full-rank | Singular matrix (can’t invert) |
| You want a least-squares solution to $Ax = b$ | Regression, ML, Optimization |
If $A$ is an $m \times n$ matrix:
Let:
$$ A = U \Sigma V^T $$Then the pseudoinverse is:
$$ A^+ = V \Sigma^+ U^T $$Where $\Sigma^+$ is obtained by:
In ML, we often solve:
$$ Ax = b \quad \text{(no exact solution if overdetermined)} $$Then the least-squares solution is:
$$ x = A^+ b $$Used in Linear Regression when $X$ is not square or full-rank.
| Use Case | Why it helps |
|---|---|
| Linear Regression | $\theta = (X^T X)^{-1} X^T y$ becomes $\theta = X^+ y$ when $X$ isn’t full-rank |
| Dimensionality Reduction | In SVD, pseudoinverse helps in low-rank approximations |
| Deep Learning | Used in layer inversion, autoencoders, or backprop through linear layers |
| Control Systems | Solve under/overdetermined linear systems |
If you're designing ML algorithms from scratch (like in research), or dealing with data where features >> samples (underdetermined), pseudoinverse gives you stable, analytical solutions — no need for iterative solvers.
++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++ ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
The Kronecker product is an operation on two matrices that produces a block matrix. It’s not element-wise, and it’s not the dot product.
For matrices $A \in \mathbb{R}^{m \times n}$ and $B \in \mathbb{R}^{p \times q}$, their Kronecker product $A \otimes B$ is of size $(mp \times nq)$
Let:
$$ A = \begin{bmatrix} a & b \\ c & d \end{bmatrix}, \quad B = \begin{bmatrix} x & y \\ z & w \end{bmatrix} $$Then:
$$ A \otimes B = \begin{bmatrix} aB & bB \\ cB & dB \end{bmatrix} = \begin{bmatrix} a x & a y & b x & b y \\ a z & a w & b z & b w \\ c x & c y & d x & d y \\ c z & c w & d z & d w \\ \end{bmatrix} $$The Hadamard product is an element-wise multiplication between two matrices (or vectors) of the same shape.
If $A, B \in \mathbb{R}^{m \times n}$, then:
$$ A \circ B = [a_{ij} \cdot b_{ij}] $$
🔁 Every element in position $(i, j)$ of the result is the product of $a_{ij} \cdot b_{ij}$.
A * B (in NumPy, PyTorch, TensorFlow when using element-wise ops)Let:
$$ A = \begin{bmatrix} 1 & 2 \\ 3 & 4 \end{bmatrix}, \quad B = \begin{bmatrix} 5 & 6 \\ 7 & 8 \end{bmatrix} $$Then:
$$ A \circ B = \begin{bmatrix} 1 \cdot 5 & 2 \cdot 6 \\ 3 \cdot 7 & 4 \cdot 8 \end{bmatrix} = \begin{bmatrix} 5 & 12 \\ 21 & 32 \end{bmatrix} $$| Use Case | Use |
|---|---|
| Element-wise operations in ML (e.g., attention, masking) | ✅ Hadamard |
| Creating structured large matrices from small ones | ✅ Kronecker |
| Efficient parameter sharing in deep learning (e.g., tensor compression) | ✅ Kronecker |
Definition/Explanation: A tensor is a generalized array with arbitrary dimensions, extending scalars (0D), vectors (1D), and matrices (2D) to higher dimensions.
In AI, tensors store complex data like images or videos.
Examples:
Why It Matters:
Definition: An ordered array of numbers (elements), e.g.
$$ \vec{v} = \begin{bmatrix} 2 \\ -1 \\ 3 \end{bmatrix} \in \mathbb{R}^3 $$
Operations:
Norm (length):
$$ \|\vec{v}\| = \sqrt{v_1^2 + v_2^2 + \dots + v_n^2} $$
A vector space is a set of vectors that can be added together and multiplied by scalars, and still remain in the set.
Formal Requirements (8 Axioms):
Let $V$ be a vector space over a field $F$ (like ℝ or ℂ). For all $\vec{u}, \vec{v}, \vec{w} \in V$, and $a, b \in F$, the following must hold:
Examples of Vector Spaces
| Space | Description |
|---|---|
| $\mathbb{R}^n$ | n-dimensional real vectors |
| Matrices | All $m \times n$ real matrices |
| Polynomials | Polynomials of degree ≤ n |
| Functions | All continuous real functions |
A subspace is a subset of a vector space that is also a vector space under the same operations.
Requirements:
For $W \subseteq V$, $W$ is a subspace if:
Example:
Word Embeddings & NLP (Natural Language Processing)
Vector Space: Words are converted into vectors in a high-dimensional vector space (like 300 dimensions in Word2Vec or 768 in BERT).
Why vector spaces? Each word is a point (vector) in this space. Words with similar meanings cluster close to each other — that’s the geometry of meaning.
Subspace idea: When you focus on specific topics (say sports-related words), you’re essentially looking at a subspace of the whole embedding space.
Real-world: Chatbots, search engines, language translation — all use vector spaces to understand semantic similarity and context.
Face Recognition Systems
What happens? Images of faces are converted into vectors (e.g., using deep learning embeddings).
Subspace models: Techniques like Eigenfaces represent face images as points in a face subspace. Recognition happens by comparing projections in this subspace.
Why This Matters for You:
Understanding vector spaces and subspaces means you can:
A linear combination is an expression of the form:
$$ a_1\vec{v}_1 + a_2\vec{v}_2 + \dots + a_n\vec{v}_n $$Where $a_i$ are scalars and $\vec{v}_i$ are vectors.
The span of a set of vectors is all possible linear combinations of those vectors.
$$ \text{Span}(\{\vec{v}_1, \vec{v}_2\}) = \{a_1\vec{v}_1 + a_2\vec{v}_2 \mid a_1, a_2 \in \mathbb{R} \} $$Example:
$$ \text{span}\left\{ \begin{bmatrix} 1 \\ 0 \end{bmatrix}, \begin{bmatrix} 0 \\ 1 \end{bmatrix} \right\} = \mathbb{R}^2 $$
If you span multiple vectors in $\mathbb{R}^n$, the result is a subspace of $\mathbb{R}^n$.
A set of vectors is linearly independent if no vector in the set can be written as a linear combination of the others.
Mathematically:
$$ a_1\vec{v}_1 + a_2\vec{v}_2 + \dots + a_n\vec{v}_n = \vec{0} \Rightarrow a_1 = a_2 = \dots = a_n = 0 $$A basis of a vector space is a set of linearly independent vectors that span the entire space.
Example:
Projection of $\vec{a}$ onto $\vec{b}$:
$$ \text{proj}_{\vec{b}} \vec{a} = \frac{\vec{a} \cdot \vec{b}}{\|\vec{b}\|^2} \vec{b} $$
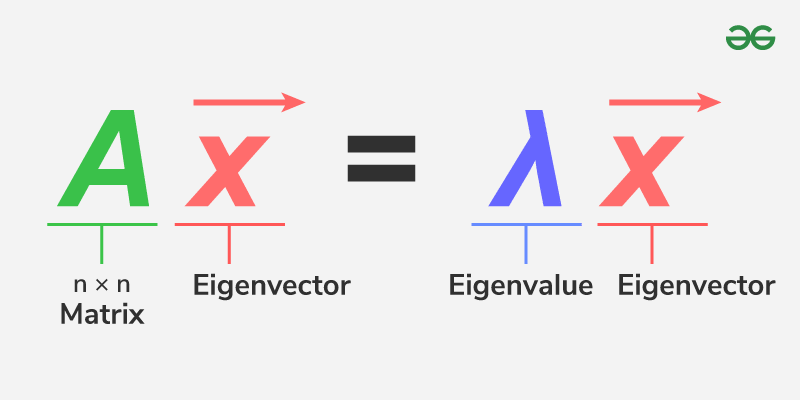
Find eigenvalues by solving the characteristic equation:
$$ \det(A - \lambda I) = 0 $$
Find eigenvectors for each $\lambda$ by solving:
$$ (A - \lambda I)\vec{v} = 0 $$
Example
$$ A = \begin{bmatrix} 4 & 1 \\ 2 & 3 \end{bmatrix} $$So eigenvalues: $\lambda_1 = 5, \lambda_2 = 2$
Eigenvector $\mathbf{v}_1 = \begin{bmatrix} 1 \\ 1 \end{bmatrix}$ (up to scalar multiples)
Properties:
Principal Component Analysis (PCA)
Applications in ML/AI
| Concept | Application |
|---|---|
| PCA (Principal Component Analysis) | Find directions (eigenvectors) of max variance |
| Spectral clustering | Eigenvectors of graph Laplacian for clustering |
| Stability analysis | Eigenvalues determine system behavior |
| Markov chains | Eigenvalues define steady states |
| Neural Networks | Eigenvalues relate to Hessian matrix in optimization |
If $A$ has $n$ linearly independent eigenvectors:
$$ A = V \Lambda V^{-1} $$What Is the Spectral Theorem?
The Spectral Theorem states that any real symmetric matrix can be diagonalized by an orthogonal matrix.
Formally:
If $A \in \mathbb{R}^{n \times n}$ is real symmetric (i.e., $A^T = A$), then:
$$ A = Q \Lambda Q^T $$Where:
Intuition
Imagine a real symmetric matrix as a "nice" transformation — like stretching or compressing along specific axes. The Spectral Theorem tells us:
There exists a special coordinate system (formed by the eigenvectors) in which the matrix just scales (not rotates or shears).
A linear transformation is a function $T: \mathbb{R}^n \to \mathbb{R}^m$ that satisfies two key properties:
Additivity:
$$ T(\vec{u} + \vec{v}) = T(\vec{u}) + T(\vec{v}) $$
Homogeneity (scalar multiplication):
$$ T(c\vec{v}) = cT(\vec{v}) $$
A transformation is linear if and only if it preserves vector addition and scalar multiplication.
Every linear transformation can be represented as matrix multiplication:
$$ T(\vec{x}) = A \vec{x} $$| Transformation | Matrix $A$ | Effect |
|---|---|---|
| Identity | $I = \begin{bmatrix} 1 & 0 \\ 0 & 1 \end{bmatrix}$ | Leaves vector unchanged |
| Scaling | $\begin{bmatrix} s & 0 \\ 0 & s \end{bmatrix}$ | Enlarges or shrinks vectors |
| Rotation (2D) | $\begin{bmatrix} \cos\theta & -\sin\theta \\ \sin\theta & \cos\theta \end{bmatrix}$ | Rotates vector by $\theta$ |
| Reflection (about x) | $\begin{bmatrix} 1 & 0 \\ 0 & -1 \end{bmatrix}$ | Flips over x-axis |
| Projection onto x-axis | $\begin{bmatrix} 1 & 0 \\ 0 & 0 \end{bmatrix}$ | Projects onto horizontal line |
Kernel (null space): Set of vectors that map to the zero vector:
$$ \ker(T) = \{ \vec{x} : T(\vec{x}) = \vec{0} \} $$
Image (range): All vectors that are outputs of $T$:
$$ \text{Im}(T) = \{ T(\vec{x}) : \vec{x} \in \mathbb{R}^n \} $$
| Property | Meaning |
|---|---|
| Linearity | Preserves addition and scalar multiplication |
| Composable | $T_1(T_2(\vec{x})) = (T_1 \circ T_2)(\vec{x})$ |
| Invertible | Exists $T^{-1}$ such that $T^{-1}(T(\vec{x})) = \vec{x}$ |
| Determined by action on basis | Knowing $T(\vec{e}_i)$ is enough to define $T$ |
| Property | Holds If... |
|---|---|
| One-to-One | $\text{ker}(A) = \{\mathbf{0}\}$ |
| Onto | Columns of $A$ span $\mathbb{R}^m$ |
| Invertible | $A$ is square and full-rank (no zero eigenvalues) |
| Use Case | Description |
|---|---|
| PCA | Projects data to directions of max variance |
| Feature transformation | Linear mappings in neural networks |
| Projections | Reduce dimensions while preserving structure |
| Affine transformations | Linear + translation used in computer vision |
For real vectors $\vec{u}, \vec{v} \in \mathbb{R}^n$:
$$ \langle \vec{u}, \vec{v} \rangle = \sum_{i=1}^{n} u_i v_i $$
An inner product space is a vector space $V$ equipped with an inner product:
$$ \langle \vec{u}, \vec{v} \rangle $$that returns a scalar and satisfies specific properties.
An inner product must satisfy:
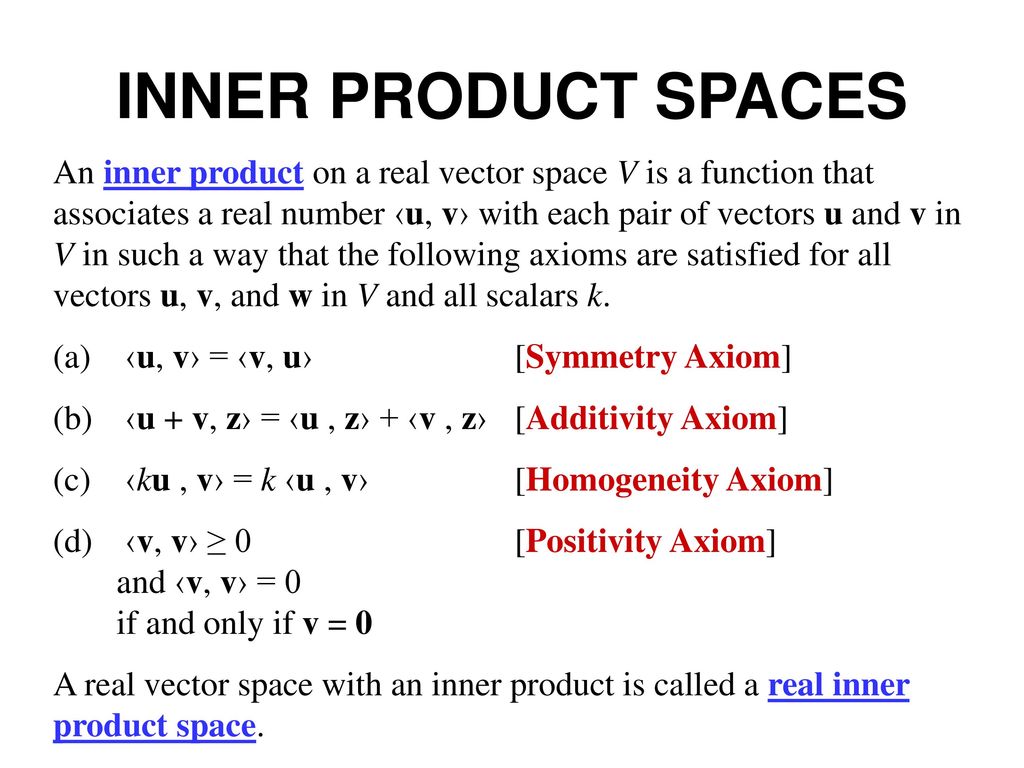
The norm (length) of a vector:
$$ \|\vec{v}\| = \sqrt{\langle \vec{v}, \vec{v} \rangle} $$Vectors $\vec{u}$, $\vec{v}$ are orthogonal if:
$$ \langle \vec{u}, \vec{v} \rangle = 0 $$Defines geometric notions of angle in abstract vector spaces.
Projection of $\vec{u}$ onto $\vec{v}$:
$$ \text{proj}_{\vec{v}} \vec{u} = \frac{\langle \vec{u}, \vec{v} \rangle}{\langle \vec{v}, \vec{v} \rangle} \vec{v} $$| Space | Inner Product Formula |
|---|---|
| $\mathbb{R}^n$ | $\sum u_i v_i$ |
| Complex vector space | $\sum u_i \overline{v_i}$ |
| Function space (e.g. $L^2$) | $\langle f, g \rangle = \int_a^b f(x)g(x)\, dx$ |
| Application | Role of Inner Product |
|---|---|
| PCA / SVD | Finding directions with high variance (via orthogonality) |
| Kernel methods (SVM) | Generalized inner products via kernel trick |
| Cosine similarity | Uses normalized inner product for text/image comparison |
| Orthogonalization | Gram-Schmidt in inner product spaces |
This means they are perpendicular in Euclidean space.
For vectors in $\mathbb{R}^n$:
$$ \vec{a} \cdot \vec{b} = \sum_{i=1}^{n} a_i b_i $$
Vectors are orthonormal if:
Common in:
Why it matters?
| Application | Use of Orthogonality & Orthonormality |
|---|---|
| PCA | Principal components are orthogonal |
| Fourier Transform | Basis functions are orthonormal |
| Gram-Schmidt | Converts a basis into orthonormal basis |
| Neural Networks | Weight initialization (orthogonal init) |
| QR Decomposition | $A = QR$, $Q$ is orthonormal matrix |
| SVD | $U$ and $V$ matrices are orthonormal |
Properties:
To project vector $\vec{a}$ onto vector $\vec{b}$:
$$ \text{proj}_{\vec{b}} \vec{a} = \frac{\vec{a} \cdot \vec{b}}{\|\vec{b}\|^2} \vec{b} $$Given a set of linearly independent vectors $\{ \mathbf{a}_1, \mathbf{a}_2, ..., \mathbf{a}_n \}$, we want to compute an orthonormal basis $\{ \mathbf{q}_1, \mathbf{q}_2, ..., \mathbf{q}_n \}$.
Steps: Projection → Subtraction → Normalization
Start with $\mathbf{u}_1 = \mathbf{a}_1$
For $k = 2$ to $n$:
$$ \mathbf{u}_k = \mathbf{a}_k - \sum_{j=1}^{k-1} \text{proj}_{\mathbf{u}_j}(\mathbf{a}_k) $$
Where projection:
$$ \text{proj}_{\mathbf{u}_j}(\mathbf{a}_k) = \frac{\mathbf{a}_k \cdot \mathbf{u}_j}{\mathbf{u}_j \cdot \mathbf{u}_j} \mathbf{u}_j $$
Normalize:
$$ \mathbf{q}_k = \frac{\mathbf{u}_k}{\|\mathbf{u}_k\|} $$
Example
Let’s take 2 vectors:
$$ \mathbf{a}_1 = \begin{bmatrix} 3 \\ 1 \end{bmatrix}, \quad \mathbf{a}_2 = \begin{bmatrix} 2 \\ 2 \end{bmatrix} $$Steps: Projection → Subtraction → Normalization
Step 1:
$$ \mathbf{u}_1 = \mathbf{a}_1 = \begin{bmatrix} 3 \\ 1 \end{bmatrix} $$Step 2:
$$ \text{proj}_{\mathbf{u}_1}(\mathbf{a}_2) = \frac{\mathbf{a}_2 \cdot \mathbf{u}_1}{\mathbf{u}_1 \cdot \mathbf{u}_1} \mathbf{u}_1 = \frac{(2)(3) + (2)(1)}{(3)^2 + (1)^2} \mathbf{u}_1 = \frac{8}{10} \mathbf{u}_1 = 0.8 \cdot \begin{bmatrix} 3 \\ 1 \end{bmatrix} = \begin{bmatrix} 2.4 \\ 0.8 \end{bmatrix} $$$$ \mathbf{u}_2 = \mathbf{a}_2 - \text{proj} = \begin{bmatrix} 2 \\ 2 \end{bmatrix} - \begin{bmatrix} 2.4 \\ 0.8 \end{bmatrix} = \begin{bmatrix} -0.4 \\ 1.2 \end{bmatrix} $$Step 3: Normalize
$$ \mathbf{q}_1 = \frac{\mathbf{u}_1}{\|\mathbf{u}_1\|} = \frac{1}{\sqrt{10}} \begin{bmatrix} 3 \\ 1 \end{bmatrix}, \quad \mathbf{q}_2 = \frac{\mathbf{u}_2}{\|\mathbf{u}_2\|} = \frac{1}{\sqrt{1.6}} \begin{bmatrix} -0.4 \\ 1.2 \end{bmatrix} $$Now $\mathbf{q}_1$ and $\mathbf{q}_2$ are orthonormal!
import numpy as np
def gram_schmidt(vectors):
orthonormal_set = []
for v in vectors:
w = v.copy()
for u in orthonormal_set:
proj = np.dot(w, u) * u
w = w - proj
norm = np.linalg.norm(w)
if norm == 0:
continue
orthonormal_set.append(w / norm)
return np.array(orthonormal_set)
# Example
a1 = np.array([3, 1])
a2 = np.array([2, 2])
vectors = [a1, a2]
Q = gram_schmidt(vectors)
print("Orthonormal basis:")
print(Q)
The covariance matrix is a square matrix that provides the covariance between pairs of variables in a dataset.
For a dataset with $n$ variables, the covariance matrix $\Sigma$ is defined as:
$$ \Sigma = \text{cov}(\vec{X}) = \frac{1}{N-1} \sum_{i=1}^{N} (\vec{x}_i - \bar{\vec{x}})(\vec{x}_i - \bar{\vec{x}})^T $$Where:
The covariance between two variables $X$ and $Y$ is:
$$ \text{cov}(X, Y) = \frac{1}{N-1} \sum_{i=1}^{N} (x_i - \bar{x})(y_i - \bar{y}) $$| Property | Description |
|---|---|
| Symmetry | Covariance matrix is symmetric: $\Sigma = \Sigma^T$. |
| Diagonal elements | Represent the variance of each variable. |
| Off-diagonal elements | Represent the covariance between pairs of variables. |
| Positive semi-definiteness | Covariance matrix is always positive semi-definite. |
| Units | Covariance is in the units of the product of the two variables. |
The correlation matrix is a normalized version of the covariance matrix, where each element is divided by the product of the standard deviations of the corresponding variables.
Where:
The correlation matrix $R$ is derived by normalizing the covariance matrix:
$$ R = \text{corr}(\vec{X}) = D^{-1} \Sigma D^{-1} $$Where:
| Property | Description |
|---|---|
| Symmetry | Correlation matrix is symmetric: $R = R^T$. |
| Range | The values of the correlation coefficient range between -1 and 1: |
| Aspect | Covariance Matrix | Correlation Matrix |
|---|---|---|
| Scaling | Depends on the units of the variables. | Unit-less (scaled). |
| Range of values | Can take any value between $-\infty$ and $\infty$. | Values range from -1 to 1. |
| Interpretation | Direct measure of joint variability. | Normalized measure of linear relationship. |
| Use | Useful for understanding the variance-covariance structure. | Useful for comparing the strength of linear relationships across different pairs. |
Given a matrix of data with 3 variables and 4 samples:
$$ X = \begin{bmatrix} 2 & 4 & 3 \\ 4 & 5 & 6 \\ 3 & 7 & 8 \\ 6 & 8 & 9 \end{bmatrix} $$Step 1: Compute the mean of each column:
Step 2: Compute the covariance between each pair of variables.
Step 3: Compute the correlation matrix by dividing each covariance by the product of the corresponding standard deviations.
| Application | Description |
|---|---|
| PCA (Principal Component Analysis) | Uses covariance matrix to find directions of maximum variance. |
| Portfolio Optimization | Correlation between assets is used to diversify risk. |
| Multivariate Analysis | Analyzing relationships and dependencies between multiple variables. |
| Regression Analysis | Correlation and covariance used to evaluate predictor variables. |
| Machine Learning | Feature selection and dimensionality reduction based on correlation. |
import numpy as np
# Example data matrix (3 variables, 4 observations)
X = np.array([[2, 4, 3],
[4, 5, 6],
[3, 7, 8],
[6, 8, 9]])
# Compute Covariance Matrix
cov_matrix = np.cov(X, rowvar=False)
# Compute Correlation Matrix
corr_matrix = np.corrcoef(X, rowvar=False)
print("Covariance Matrix:\n", cov_matrix)
print("Correlation Matrix:\n", corr_matrix)
Covariance Matrix: [[2.91666667 2.33333333 3.5 ] [2.33333333 3.33333333 4.66666667] [3.5 4.66666667 7. ]] Correlation Matrix: [[1. 0.74833148 0.77459667] [0.74833148 1. 0.96609178] [0.77459667 0.96609178 1. ]]
Matrix factorization is the process of decomposing a matrix $A \in \mathbb{R}^{m \times n}$ into two (or more) matrices whose product approximates the original matrix.
$$ A \approx B \cdot C $$Where:
Singular Value Decomposition (SVD)
Decomposes a matrix $A \in \mathbb{R}^{m \times n}$ into three matrices:
$$ A = U \Sigma V^T $$
Non-negative Matrix Factorization (NMF)
Decomposes $A$ into two matrices $W \in \mathbb{R}^{m \times k}$ and $H \in \mathbb{R}^{k \times n}$, where all elements are non-negative:
$$ A \approx W H $$
LU Decomposition
Factorizes a square matrix $A$ into:
$$ A = L U $$
QR Decomposition
Decomposes $A$ into:
$$ A = Q R $$
Cholesky Decomposition
For a positive-definite matrix $A$, it is decomposed into:
$$ A = L L^T $$
| Use Case | Technique | Description |
|---|---|---|
| Recommendation Systems | SVD, NMF | Factorizes user-item interaction matrix to predict ratings |
| Dimensionality Reduction | SVD, PCA | Reduce dimensionality while preserving variance |
| Topic Modeling | NMF | Factorizes text data into topics, each represented as a combination of words |
| Image Compression | SVD, NMF | Decomposes image matrix to reduce storage while preserving features |
| Signal Processing | NMF, SVD | Decomposes signals into components for analysis or denoising |
| Data Compression | SVD, NMF | Reduces data size while retaining most important features |
| Property | Description |
|---|---|
| Low-rank approximation | Factorization approximates matrix using fewer components |
| Sparsity | NMF typically enforces sparsity (non-negative elements) |
| Uniqueness | In general, matrix factorization does not yield a unique solution without constraints |
| Computational Complexity | Factorization techniques like SVD are computationally expensive (especially for large matrices) |
| Interpretability | Factorized matrices (especially NMF) are often easier to interpret (e.g., topics, latent features) |
Dimensionality reduction: Use the top $k$ singular values and vectors to approximate $A$.
$$ A_k \approx U_k \Sigma_k V_k^T $$
Objective: Find matrices $W$ and $H$ that minimize the Frobenius norm:
$$ \| A - WH \|_F $$
(i.e., the difference between the original matrix and the approximation).
Use gradient descent or alternating least squares (ALS) methods for optimization.
| Factorization Method | Best Use Case | Key Advantage | Limitation |
|---|---|---|---|
| SVD | Dimensionality reduction, PCA | Provides exact decomposition | Computationally expensive for large matrices |
| NMF | Text mining, topic modeling | Interpretability (non-negative factors) | Can only handle non-negative data |
| LU | Solving systems of linear equations | Fast for square matrices | Requires square matrices |
| QR | Solving least-squares problems | Computationally stable | Not ideal for large-scale systems |
import numpy as np
# Example matrix A
A = np.array([[1, 2, 3], [4, 5, 6], [7, 8, 9]])
# Perform Singular Value Decomposition (SVD)
U, Sigma, Vt = np.linalg.svd(A)
# Reconstruct the matrix
A_reconstructed = np.dot(U, np.dot(np.diag(Sigma), Vt))
print("Original Matrix:\n", A)
print("Reconstructed Matrix:\n", A_reconstructed)
Important identities:
Pythagorean:
$$ \sin^2 x + \cos^2 x = 1 $$
Angle sum/difference:
$$ \sin(a \pm b) = \sin a \cos b \pm \cos a \sin b $$
$$ \cos(a \pm b) = \cos a \cos b \mp \sin a \sin b $$
Double angle:
$$ \sin 2x = 2 \sin x \cos x, \quad \cos 2x = \cos^2 x - \sin^2 x $$

General steps:
Example 1: Solve for $x$ in $\sin x = \frac{1}{2}$ on $[0, 2\pi]$
Example 2: Solve $2 \cos^2 x - 3 \sin x = 0$
Form:
$$ f(x) = a^x, \quad a > 0, a \neq 1 $$
Properties:
Natural exponential:
$$ e^x = \lim_{n \to \infty} \left(1 + \frac{x}{n}\right)^n, \quad e \approx 2.718 $$
Inverse of exponential:
$$ y = \log_a x \iff a^y = x $$
Properties:
Change of base formula:
$$ \log_a x = \frac{\log_b x}{\log_b a} $$
Common logarithms:

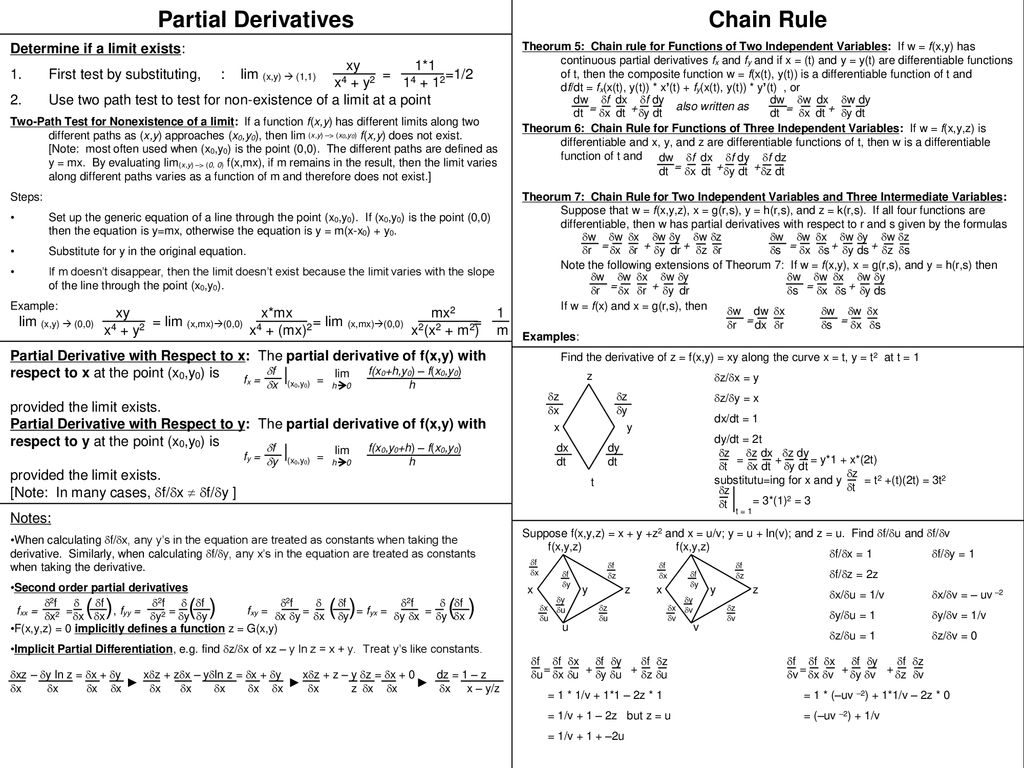
For scalar function $f(x, y, z)$:
$$ \nabla f = \left( \frac{\partial f}{\partial x}, \frac{\partial f}{\partial y}, \frac{\partial f}{\partial z} \right) $$For vector field $\vec{F} = (F_1, F_2, F_3)$:
$$ \nabla \cdot \vec{F} = \frac{\partial F_1}{\partial x} + \frac{\partial F_2}{\partial y} + \frac{\partial F_3}{\partial z} $$For scalar function $f(x, y, z)$:
$$ \nabla^2 f = \frac{\partial^2 f}{\partial x^2} + \frac{\partial^2 f}{\partial y^2} + \frac{\partial^2 f}{\partial z^2} $$For scalar field $f$ over curve $C$:
$$ \int_C f \, ds $$For vector field $\vec{F}$ over path $C$:
$$ \int_C \vec{F} \cdot d\vec{r} $$For scalar field:
$$ \iint_S f(x, y, z) \, dS $$For vector field $\vec{F}$:
$$ \iint_S \vec{F} \cdot \vec{n} \, dS $$Used when transforming multivariate functions.
Let $\vec{f}(\vec{x}) = [f_1(x), f_2(x), ..., f_m(x)]^\top$, where $\vec{x} = [x_1, x_2, ..., x_n]^\top$
Then the Jacobian $J \in \mathbb{R}^{m \times n}$ is:
$$ J = \frac{\partial \vec{f}}{\partial \vec{x}} = \begin{bmatrix} \frac{\partial f_1}{\partial x_1} & \cdots & \frac{\partial f_1}{\partial x_n} \\ \vdots & \ddots & \vdots \\ \frac{\partial f_m}{\partial x_1} & \cdots & \frac{\partial f_m}{\partial x_n} \end{bmatrix} $$✅ Used in:
Used for second-order optimization analysis (e.g., Newton’s method).
Let $f: \mathbb{R}^n \to \mathbb{R}$
Then the Hessian $H \in \mathbb{R}^{n \times n}$ is:
$$ H = \nabla^2 f = \begin{bmatrix} \frac{\partial^2 f}{\partial x_1^2} & \cdots & \frac{\partial^2 f}{\partial x_1 \partial x_n} \\ \vdots & \ddots & \vdots \\ \frac{\partial^2 f}{\partial x_n \partial x_1} & \cdots & \frac{\partial^2 f}{\partial x_n^2} \end{bmatrix} $$✅ Used in:
Optimization algorithms are at the heart of machine learning and deep learning. They are used to minimize (or maximize) a loss (or objective) function by iteratively updating the model's parameters.
Objective Function: The objective or the loss function measures how well your model is performing. It quantifies the difference between the predicted output and the actual target.
Goal of Optimization: Minimize the loss function.
Variables: The following are the Parameters or the internal variables of the model that are learned during training (e.g., weights in a neural network).